Multiomics
Correlations between different data types
We plan to integrate the analysis of datasets from different types of assays. We think this will yield additional insights into the pathophysiology of ME/CFS and in particular, point toward molecular mechanisms of PEM. Integrating our multi-omic data with the clinical and exercise physiology data of our patient cohort may allow us to identify unique molecular signatures in different subsets of ME/CFS patients. These molecular signatures may provide a basis for tailored diagnostics and treatments for clinical and demographic subsets of ME/CFS patients in the future.
Predictive modeling using a combination of molecules in different classes that have significantly different levels in ME/CFS patients and controls at baseline may enable more accurate classification of ME/CFS patients and healthy controls than could an assay utilizing biomarkers only from a single class.
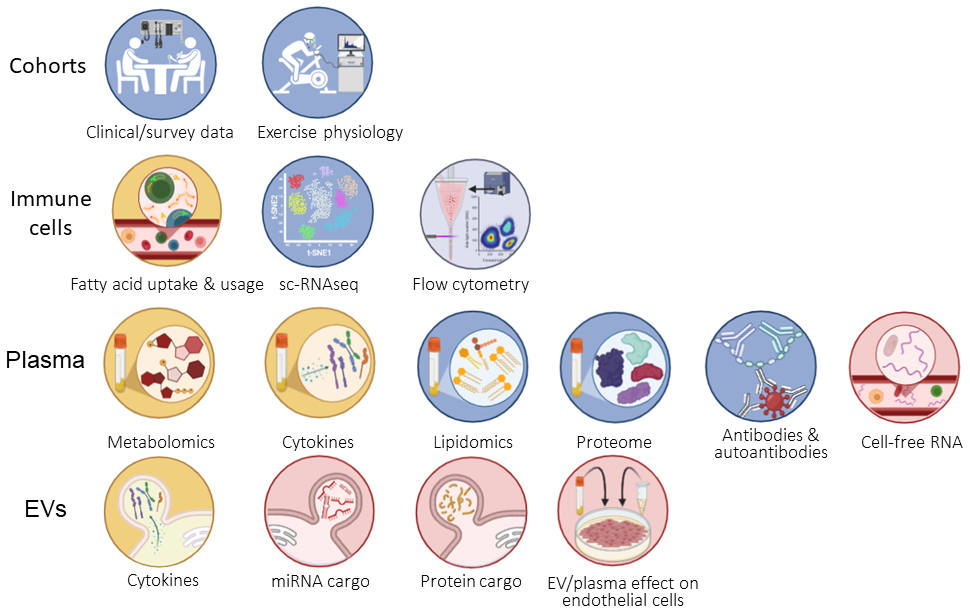
Researchers from all the projects will be involved in data comparisons, led by Dr. Katherine Glass in the Hanson lab, Andrew Grimson’s collaborator Dr. Dawei Li (Texas Tech University), and Jen Grenier’s group in the Genomic Innovation Hub (especially Dr. Paul Munn and Dr. Adrian McNairn). This work is funded by the NIH (U54AI178855).
 Center for Enervating NeuroImmune Disease
Center for Enervating NeuroImmune Disease