Differential Gene Expression in Muscle Cells
The goal is to identify molecular and cellular alterations present in ME/CFS skeletal muscles through innovative approaches to capture the transcriptome and epigenome with single-cell and spatial resolution in human biopsies. Using single nuclei isolated from muscle biopsies, the genes expressed in each cell, as well as the configuration of chromosomes in each cell, will be determined. Gene expression information will also be obtained from small regions of cross-sections (slices) of muscle tissue (spatial transcriptomics). All of this information will be used to determine whether specific types of muscle-resident cells are dysregulated at the transcriptional and epigenetic levels in ME/CFS subjects vs. controls.
Gene expression in uninfected mouse heart slices (longitudinal sections)
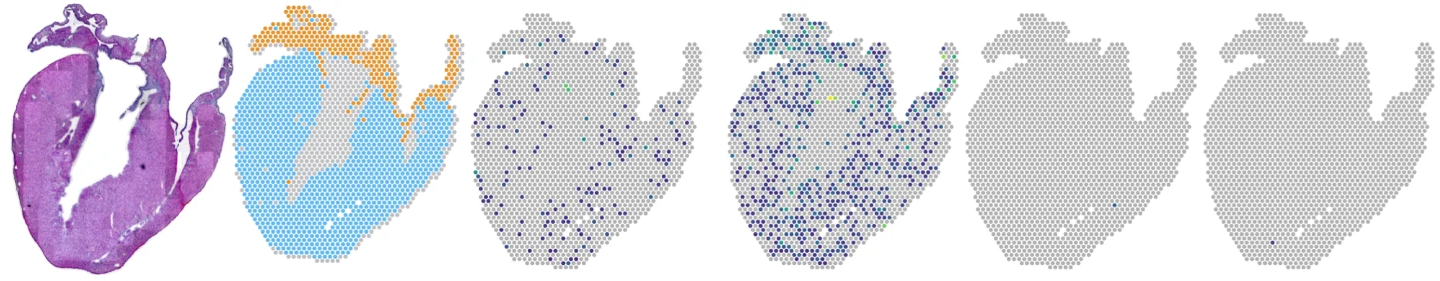
Hearts from mice infected with reovirus
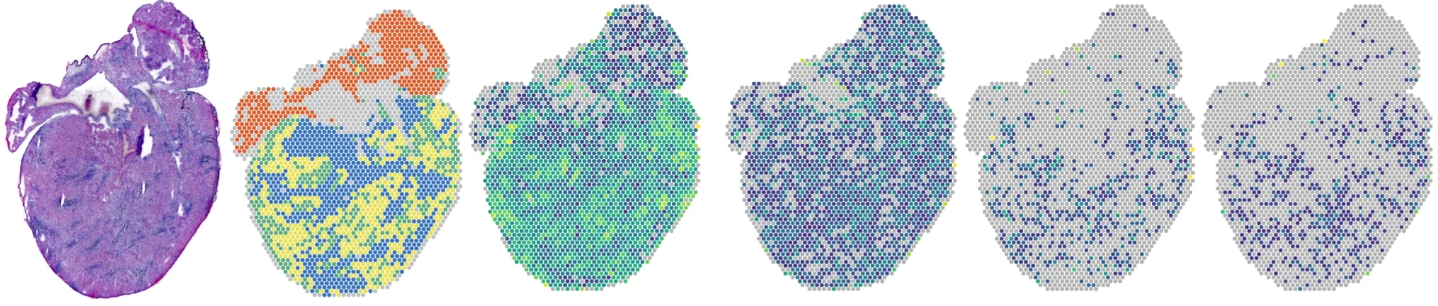
Spatial mapping of the total transcriptome by in situ polyadenylation
This NIH funded (U54AI178855) project will be led by Dr. Ben Cosgrove, who will be closely collaborating with Dr. Iwijn DeVlaminck; both are faculty in the Cornell Meinig School of Biomedical Engineering. Molecular data analysis will also occur through a collaboration with Dr. Jen Grenier, the Genomics Core Lead who heads the Cornell Genomics Innovation Hub.
 Center for Enervating NeuroImmune Disease
Center for Enervating NeuroImmune Disease